As a core objective we aimed to provide a science gateway that serves as a technical foundation, enabling plant researchers to access approaches inspired by software engineering for data management. Over the past two years, we have developed the DataPLANT DataHUB, a comprehensive platform offering various research data management (RDM) workflows to support data scientists throughout the entire data life cycle. These workflows encompass activities such as data annotation, structuring, and ultimately, the publication of research outcomes. A further aim projected for the third year of DataPLANT was to enable the DataHUB to serve as the primary entry point and provide researchers with access to a range of analysis workflows. The workflow descriptions are stored in the Annotated Research Context (ARC), ensuring compatibility with processing frameworks like Galaxy and nf-core nextflow pipelines. This should allow researchers to seamlessly integrate their analysis pipelines with the DataHUB, facilitating efficient data processingand data reuse. As a first major step, we have developed an assisted workflow specifically tailored for data publication using CERN's Open Source repository software InvenioRDM. This workflow simplifies the process of sharing research data by providing guidance and support to researchers during the publication stage.
Galaxy workflow system
In DataPLANT, our data-centric approach revolves around the ARC, which serves as the central entity for all digital processes. This includes data processing and analysis workflows, where Galaxy is meant to play a crucial role as part of the DataPLANT ecosystem. The Galaxy community is renowned as one of the largest bioinformatics communities globally, offering a vast array of over 3000 tools and a wide range of bioinformatics and data processing workflows specifically tailored for researchers in the fundamental plant research community. Galaxy's core framework provides several abstraction layers that allow for easy extension and integration with new technologies. Primarily, advanced bioinformatics software is made accessible to scientists worldwide through its intuitive web interface. Plus, Galaxy promotes reproducibility by automatically creating re-runnable protocols for each analysis, enabling researchers to reproduce and validate their results. This is about to get integrated with the DataPLANT DataHUB.
A crucial subsystem within Galaxy is the handling of user data. At the moment it supports various types of data storage, including hierarchical POSIX storage, S3, and iRODS. These storage options can be assigned to users, groups, or roles, allowing for flexible quota assignments per object store. Additionally, Galaxy provides a user-friendly system for browsing and importing public data deposited in various storage platforms such as S3, SFTP, WebDAV, or Dropbox accounts. Furthermore, Galaxy supports the export of research artifacts, including workflow invocations. Currently, these artifacts can be exported as BioCompute Objectsand RO-Crates. Here, DataPLANT is working on introducing support for Research Objects and ARCs. A first prototype of the ARCfs got integrated via a past Galaxy update. ARCfs supports read and write access and is being tested as a workflow integration in Galaxy.
Galaxy and DataPLANT DataHUB integration
The ARCfs component is designed to provide cross-site and cross-service access to ARC by offering a file-system-like view to Galaxy and potentially other services in the future. It leverages PyFilesystem2, a Python library that provides a Framework for file-system abstractions. Unlike other file-system views on Git, ARCfs communicates exclusively with GitLab's REST API to retrieve file-metadata and content, including Large File Storage (LFS) files.

Figure 1.The ARCfs file system abstraction
as intermediate layer, connecting Galaxy
and DataHUB ARCs.
Integrating ARCfs with Galaxy has several advantages. Users can easily browse and load content from all ARCs they have access to through a web interface, as shown in figure 3, eliminating the need to search for specific repositories on GitLab or other tools. Additionally, ARCfs enables direct transfer of files from GitLab to the Galaxy server, eliminating the need for manual downloading and uploading. This not only saves time but also avoids the complexity of partial repository clones when dealing with large ARCs where only a subset of files is required.
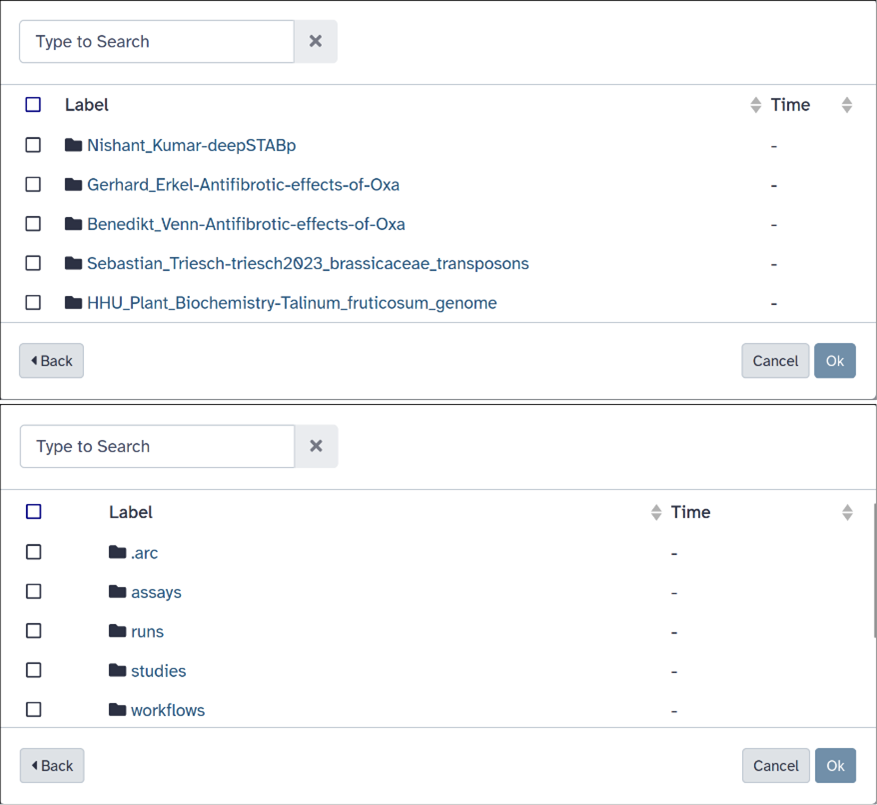
Figure 2.The ARCfs integrated in Galaxy. View over all accessible ARCs (top)
and inside view of an ARC (bottom).
ARCfs currently only supports read access and is being tested as a workflow integration in Galaxy. Accessing public ARCs hosted on DataHUB does not require any configuration, while accessing private repositories requires a GitLab access token with the appropriate scope. Furthermore, a version of ARCfs with read-write access is being discussed to allow users to write computation results from Galaxy directly back into an ARC. To write such results or other datasets into an ARC, a user will be able to browse and choose an ARC which the selected dataset should be exported to. Upon triggering this export, the dataset will be uploaded as an LFS file into a newly created branch. Subsequently, a merge request into the default branch is created. This allows users to review the datasets before they are adopted into the ARC.